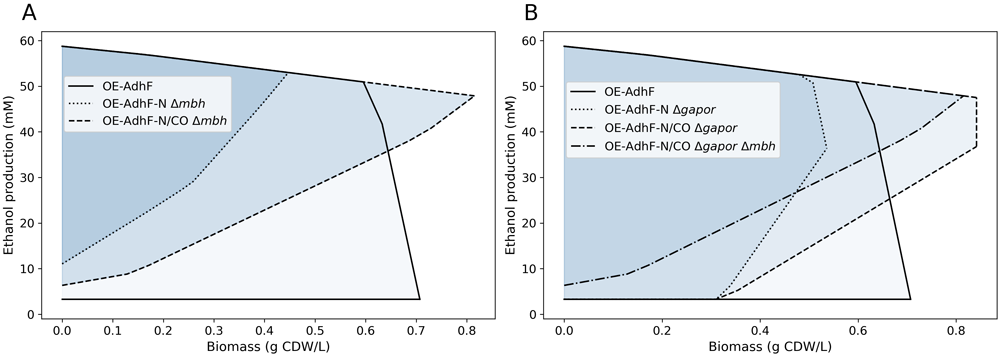
Model-informed Metabolic Engineering
The bio-based production of organic chemicals provides a sustainable alternative to fossil-based production in the
face of today’s climate challenges. Our research seeks to inform the metabolic engineering of hyperthermophilic
organisms using computational models. Two of the target organisms include the bacterium Caldicellulosiruptor
bescii and the archaeon Pyrococcus furiosus. Our ongoing research leads to the identification of new engineering
designs for increasing the production yields of bio-based chemicals.
Publications: Zhang et al., mSystems (2021); Rodionov et al., mSystems (2021); Crosby et al., AEM (2022); Lipscomb et al., AEM (2023); Vailionis et al., AEM (2023). Funding: DOE DE-SC0019391 DE-SC0022192
Community Dynamics and Succession of Microbiomes
Microbes are among the most abundant organisms on Earth and play important roles in primary production, respiration, and nutrient cycling. Besides their contributions to global biogeochemical cycles, microbes may produce molecular signals for rapidly sensing and responding to environmental changes. We combine meta-omics with computational modeling to study the genotype-to-phenotype association of microbes in their natural community. Our research led to the molecular characterization of host-associated microbiomes and marine protists. Examples of recent applications included characterizing metabolic versatility of marine protists in oxygen-depleted environments and identifying the metabolism and evolution of marine invertebrate-associated bacteria using community sequencing and metagenomics.
Publications: Pimentel et al., mSphere (2021); Gomaa et al., Science Adv. (2021); Powers et al., Frontiers in Marine Sci. (2022). Funding: NSF IOS # 1557566, 1557430, OIA # 1929078
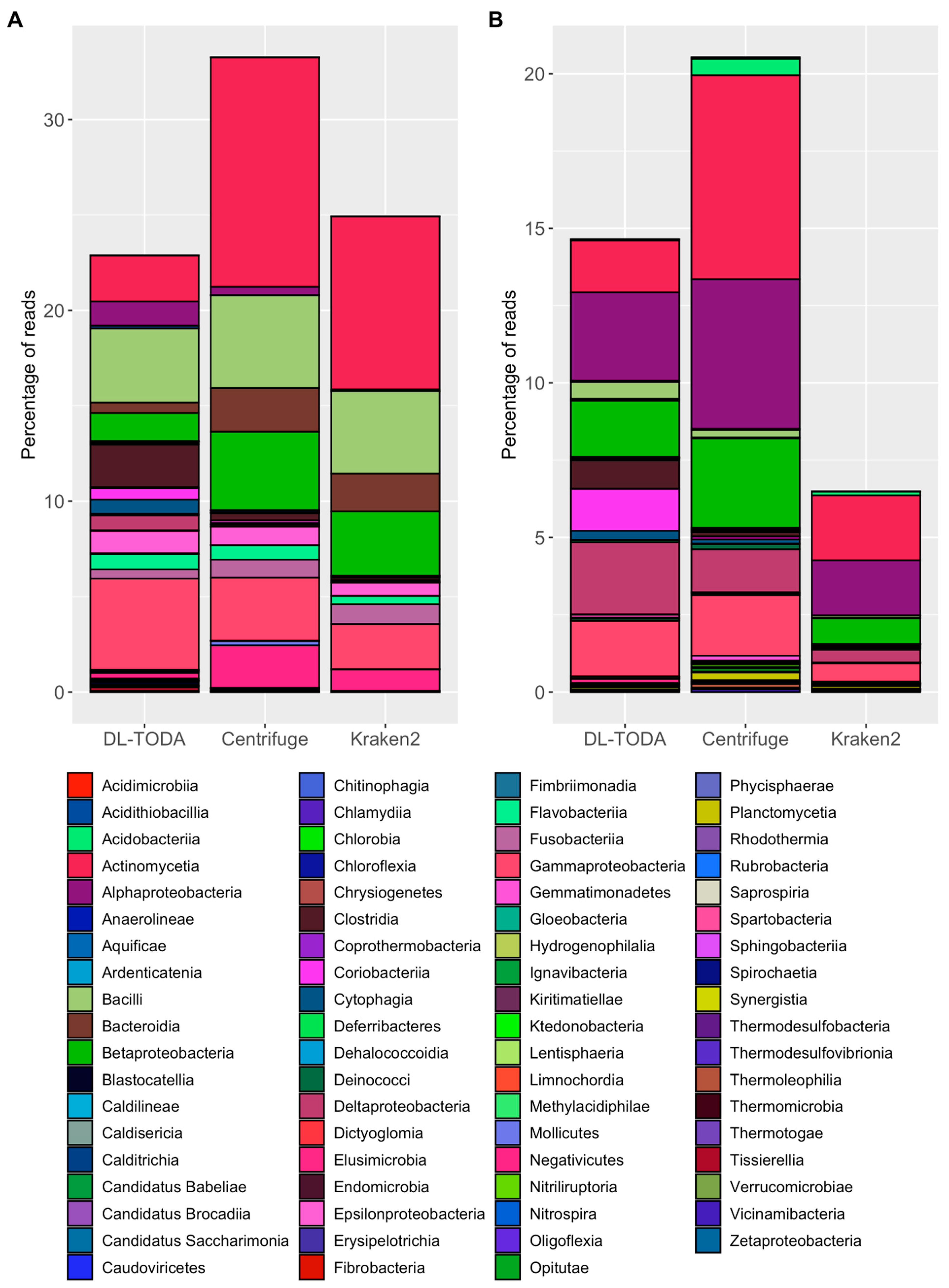
Deep Learning Tool for Omics Data Analysis
The rapid development of deep learning techniques has inspired new applications that leverage the massive molecular
data collected from a variety of environmental samples. Here we demonstrated the application of deep learning in
metagenomics, a technique frequently used for genome-wide profiling of microbiomes. Our recent release of a deep
learning model, DL-TODA ,
supports the rapid taxonomic classification of large-scale metagenomic reads with high accuracy.
Publication: Cres et al. (2023). Funding: NSF DBI # 1553211